Ten years ago, the discovery of sodium taurocholate cotransporting polypeptide (NTCP) as receptor for HBV has finally allowed to perform infection experiments in cell culture.
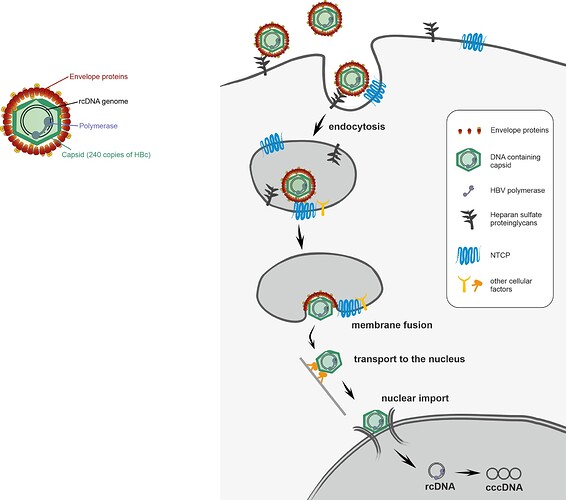
Nevertheless, many steps of how HBV enters its host cells remain unclear. HBV is known to attach on the surfaces of liver cells to heparan sulfate proteoglycans before specifically binding to the receptor NTCP. This interaction is guided by the HBV envelope proteins (specifically the L protein). It is proposed that HBV is then taken up into the cell by endocytosis, which means it is still wrapped in a cellular membrane and must fuse with this membrane to release the virus capsid into to cell. Then, the HBV capsid is transported to the nucleus which it enters through the nuclear pore complex, which controls import into and export from the nucleus. Only after entering the nucleus, HBV can establish its persistent genome and start expressing proteins and finally new viruses.
Part of our recent research has focused on the fluorescent labelling of Hepatitis B virus. This would be very helpful for microscopy studies of HBV entry and hopefully allow us to study HBV entry in more detail. For example, it remains unknown where and how membrane fusion takes place during endocytosis and whether the entire virus capsid can enter the nuclear pore complex as its size is close to the upper size limit of the pore. Ideally the fluorescent label would not affect viral functions and give a very strong signal to allow specific tracking of the virus during live microscopy.
From the start, many factors had to be considered for a labelling strategy. We had to decide which part of the virus to label. We excluded the envelope of HBV for two reasons: First, envelope labelling would not allow tracking to the nucleus but only until membrane fusion in the endosome, and second HBV produces a large amount of empty envelopes (also known as HBsAg) which would give a lot of fluorescent signal not specific for the infectious HBV particle. This left the capsid of HBV which consists of 240 copies of the core protein HBc, one copy of the HBV polymerase and the genome. Even though many fluorescent compounds for DNA labelling exist, the HBV genome is very small and probably the fluorescent signal not strong enough. Therefore, we aimed to label the capsid protein of HBV, which also comes with some difficulties. The HBV capsid is densely packed and forms multiple interactions with the envelope proteins, therefore insertion of large fluorescent tags are very likely to interfere with the assembly and function of the virus. Finally, we employed a method called genetic code expansion. As the name says, this method expands the genetic code and allows the introduction of unnatural amino acids into a protein of interest. For the purpose of HBV labelling, we use amino acids which can be specifically labelled with small chemical dyes. This combination of unnatural amino acids and synthetic dyes constitutes the smallest available fluorescent tag for specific protein labelling.
Nevertheless, this method still requires a lot of optimisations, because the incorporation of these unnatural amino acids is not very efficient, and the tight organisation of the HBV virion does not easily allow insertion even of this small fluorescent tag. As future prospective, we hope to be able to use fluorescently tagged HBV to study the dynamics and interactions of HBV entry on a cellular level.
I hope I could give you a glimpse into the basic research we are doing, and I am happy to take questions!
 .
.